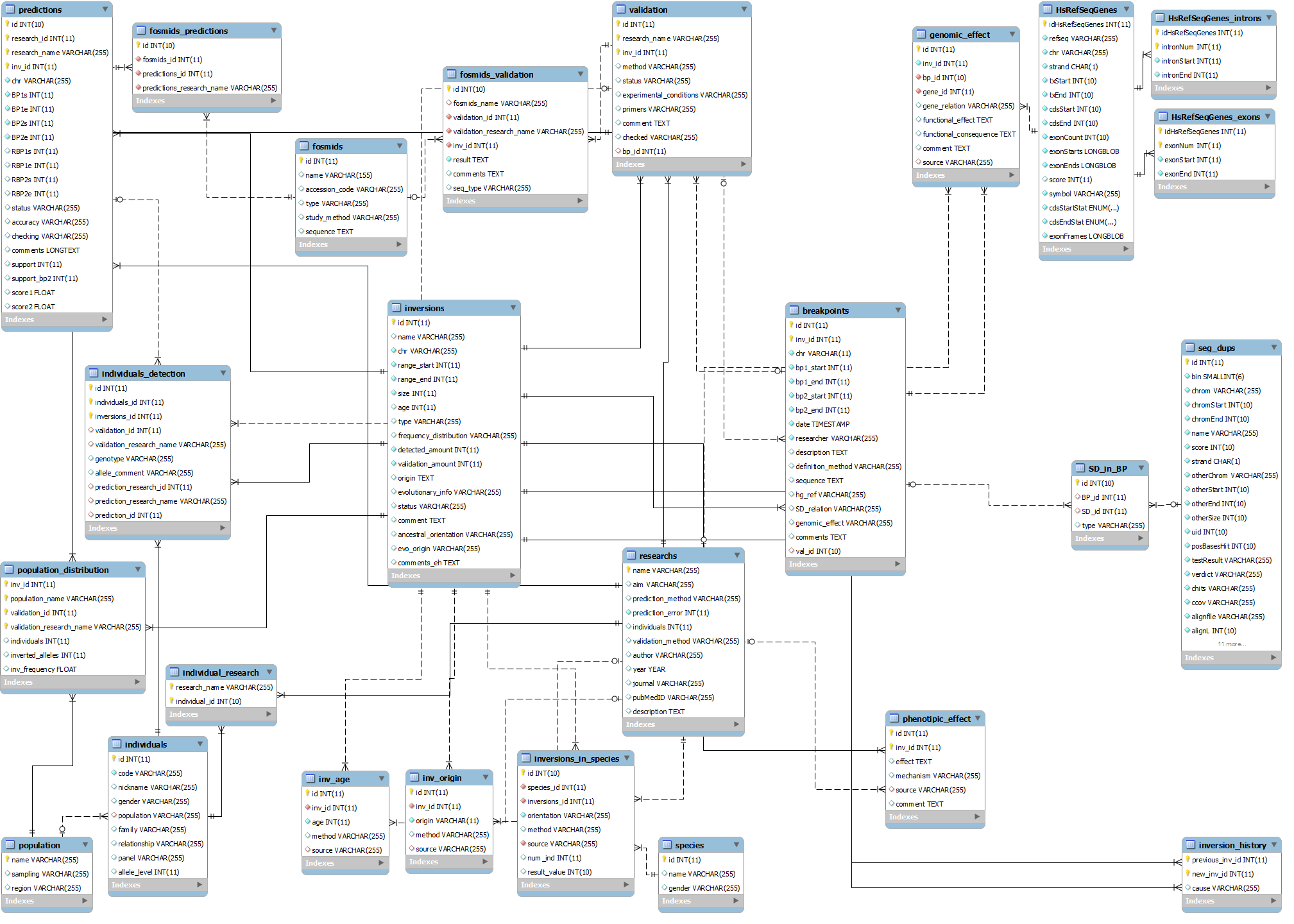
InvFEST is a data-warehouse implementation, commonly used to start schema data models. The center of the star is the “inversion” table, which is de-normalized for speed purposes. The points of the star are the dimension tables that contain related information (e.g. predictions, validations, evolutionary information, etc.). Below follows a detailed Entity-Relationship (ER) schema of InvFEST and a list enumerating all the tables of the database with a description and all relevant areas of the InvFEST processing engine they are involved in.
|
Table Name
|
Description
|
Relevant Areas of the
InvFEST
Processing Engine
|
|
breakpoints
|
Second central table of the database. Contains a summary of all basic information about all inversion breakpoints in the database (chromosome location, definition date, source, description, association with segmental duplications, association with genes, general comments, validation ID). Includes all defined breakpoints for all inversions during all the history of the database; those displayed as final breakpoints of the inversion in the inversion report are the latest manually defined breakpoints in the table if available, or the latest defined breakpoints in the table otherwise
|
Predictions
,
Validations
,
Merging Engine
|
|
fosmids
|
Stores codes and accession numbers of
fosmids
used by predictions and/or validations
|
Predictions
,
Validations
|
|
fosmids_predictions
|
Linking table between tables
predictions
and
fosmids
(which
fosmids
are supporting each prediction, if applicable)
|
Predictions
|
|
fosmids_validation
|
Linking table between tables
validation
and
fosmids
(which
fosmids
are supporting each validation, if applicable)
|
Validations
|
|
genomic_effect
|
Association with genes (
RefSeq
genes annotated on hg18) for each defined breakpoints of the database (
bioinformatic
prediction of how the breakpoints affect the gene, observed functional effects on the gene, source). Automatic associations included in the table are conservative: only shown if the gene sequence is disrupted by the inversion (not if the complete gene is inverted or is close to the inversion), and only shown if no transcripts of a gene are left unaffected; when affected, if different transcripts are affected differently, only less drastic effect is described
|
Predictions
,
Validations
,
Associated information
,
Merging Engine
|
|
HsRefSeqGenes
|
RefSeq
genes on hg18 as downloaded from the UCSC
|
Predictions
,
Merging Engine
|
|
HsRefSeqGenes_exons
|
Exons
extracted from the
HsRefSeqGenes
table
|
Predictions
,
Merging Engine
|
|
HsRefSeqGenes_introns
|
Introns
extracted from the
HsRefSeqGenes
table
|
Predictions
,
Merging Engine
|
|
individual_research
|
Individual codes analyzed in specific studies
|
Predictions
,
Validations
|
|
individuals
|
Individual codes with associated information (nickname, gender, population, family, relationship in the family, panel)
|
Associated information
|
|
individuals_detection
|
Links IDs of individuals to specific predictions and/or validations
|
Predictions
,
Validations
|
|
inv_age
|
Estimated age for inversions (source, method, estimated age)
|
Validations
,
Associated information
|
|
inv_origin
|
Evolutionary origin of inversions (source, method, unique/recurrent)
|
Validations
,
Associated information
|
|
inversion_history
|
History report of inversions manually merged or split in the database (previous inversion ID, new inversion ID, cause)
|
Validations
|
|
inversions
|
Central table of the database. Contains a summary of all basic information about all inversions in the database (name, chromosome location, size, frequency, number of predictions, number of validations, origin, status)
|
Predictions
,
Validations
,
Associated information
,
Merging Engine
|
|
inversions_in_species
|
Orientation of the inversion in species other than humans (species ID, source, method, orientation: standard=equal to hg18 reference assembly / inverted=opposite to hg18 reference assembly)
|
Validations
,
Associated information
|
|
phenotipic_effect
|
Observed phenotypic effects of inversions in the database (source, mechanism, observed effects)
|
Validations
,
Associated information
|
|
population
|
Population name and region name
|
Associated information
|
|
population_distribution
|
Stores population distribution information (number of analyzed individuals, number of inverted alleles, population frequency), for specific populations obtained by specific validations
|
Validations
,
Associated information
|
|
predictions
|
Stores all predictions from individual studies (including coordinates, status,
bioinformatic
scores), and links to tables
inversions
(there is redundancy in the
predictions
table because the relationship is
m:n
) and
researchs
|
Predictions
,
Merging Engine
|
|
researchs
|
Information about research articles from which
InvFESTdb
information is extracted (name, description, citation, number of individuals analyzed, prediction method, prediction error, validation method)
|
Predictions
,
Validations
,
Associated information
|
|
SD_in_BP
|
Association with segmental duplications (as downloaded from the UCSC) for each defined breakpoints of the database. Also describes if the same segmental duplication affects one or both breakpoints, and in the latter case, if the segmental duplication is in direct or indirect orientation in one breakpoint versus the other
|
Predictions
,
Merging Engine
|
|
seg_dups
|
Segmental duplications onhg18 as downloaded from the UCSC
|
Predictions
,
Merging Engine
|
|
species
|
List of species other than humans (ID, species common name)
|
Associated information
|
|
validation
|
Stores all validations from individual studies (including method, experimental conditions, status), and links to table
researchs
|
Validations
|